Hybrid Quantum-Classical Graph Neural Networks
Hybrid GNN-VQC model for image classification
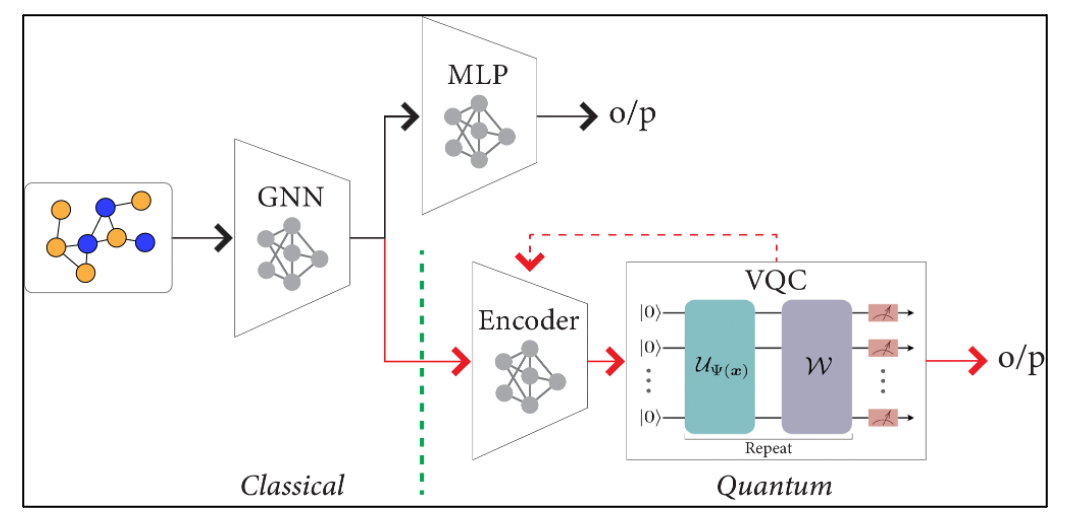
Understanding how tumor cells self-organize and interact within the tumor microenvironment (TME) is a long standing question in cancer Biology. In This project, we modelled TME as a graph structure to comprehensively encode the tissue composition in terms of biologically meaningful entities, such as cells, tissues, and their interactions. As a result, GNNs can elegantly integrate cellular information with tumor morphology, topology, and interactions among cells and/or tissue structures. We created a hybrid classical-quantum network which combines a GNN with a Variational Quantum Classifier (VQC).
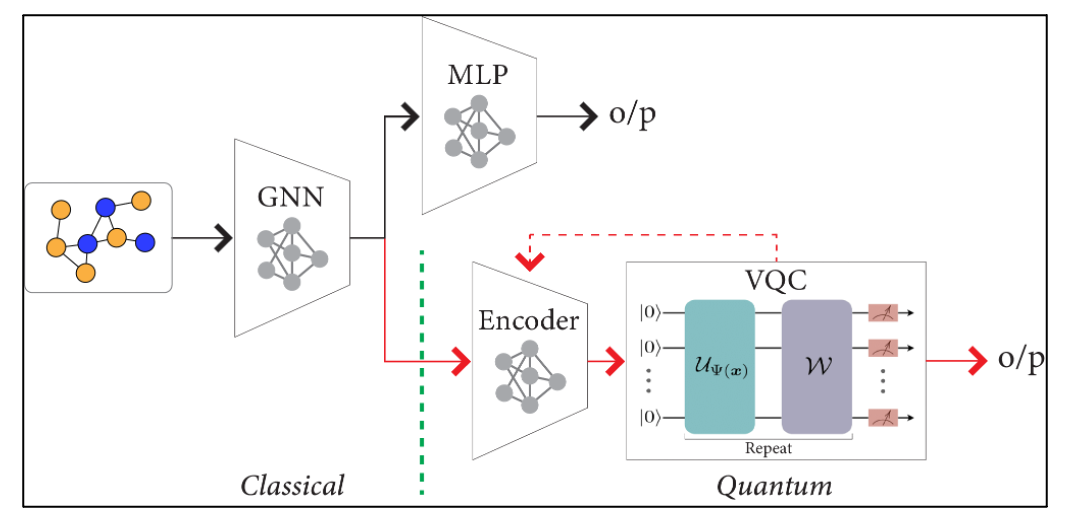
We followed two approaches for training our Hybrid Network with different encoding schemes and feature dimensions:
(i) with a pretrained GNN (having frozen weights)
(ii) with trainable GNN and VQC parameters to backpropogate simultaneously.
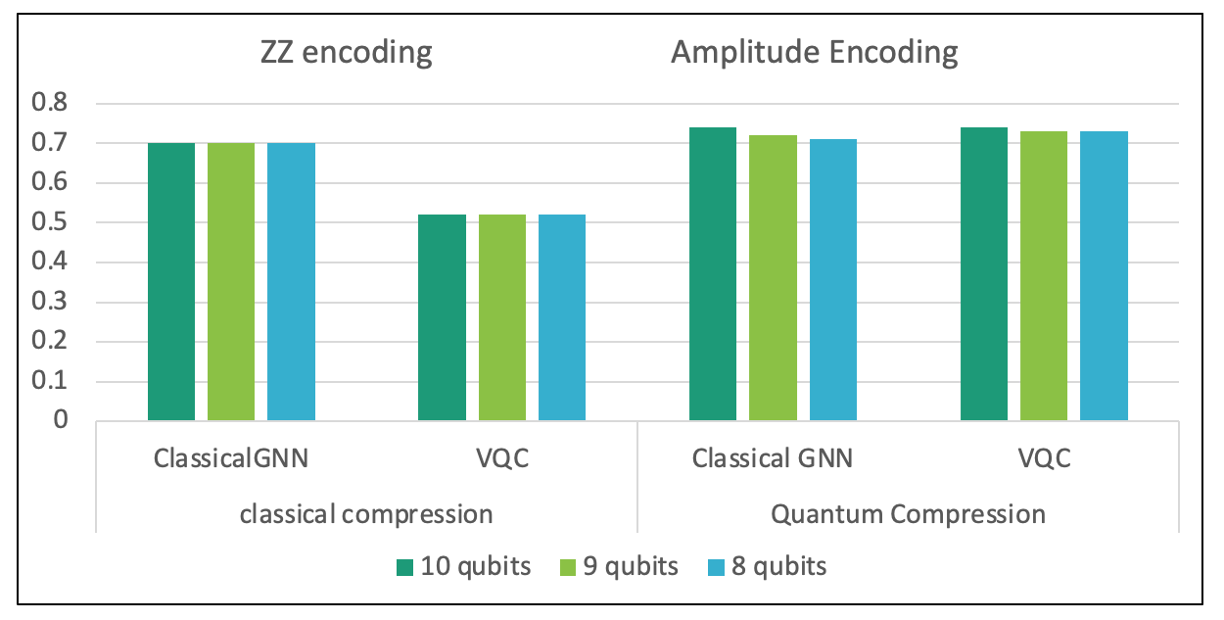
We showed that end-to-end training is significantly better than serially training such models and demonstrate results on a real-world breast-cancer subtyping task. In detailed ablation studies we observed that quantum compression can be significantly better to qubit requirements without information loss unlike lossy classical compression. Future directions could be to explore how other such classical networks can be combined with quantum circuits to enhance their trainability and improve generalization.
Source code:
References
2023
-
 Hybrid quantum-classical graph neural networks for tumor classification in digital pathology2023
Hybrid quantum-classical graph neural networks for tumor classification in digital pathology2023